
Novel Deep Learning Model For The Detection Of Cardiac Amyloidosis, A Pilot Reader Studyy
- | By Ultromics
Jeremy A. Slivnick, Ashley Ackerman, Jorge Oliveira, Will Hawkes, Agis Chartsias, Ross Upton, Federico M. Asch, Juan I. Cotella, Linda Lee, Giancarlo Saldana, Christopher Fernandez, Jia Guo, Karima Addetia, Matthew Maurer, Steven Helmke, Vidhushei Yogeswaran, Richard Cheng,
James N. Kirkpatrick, Karolina Zareba, Masaaki Takeuchi, Tetsuji Kitano, Marcelo Luz, Viviane Hotta, Aldo Prado, Pablo Ellisamburu, Marielle Scherrer-Crosbie, Marwa Soltani, Akhil Narang, Gary Woodward, Roberto M. Lang.

Background
While TTE remains the frontline imaging modality for patients with cardiac amyloidosis (CA), classical TTE findings often lack sensitivity, resulting in delayed diagnosis and treatment.
The aim of this study was to develop a deep learning (DL) model to detect CA and perform a preliminary assessment of its capabilities to assist readers.
Methods
We trained and validated (75%/25% split) a 3D convolutional neural network to detect CA using 2757 apical 4-chamber (A4C) images derived from confirmed CA patients and controls.
Utilizing a separate test dataset of 60 CA (30 AL, 30 ATTR) and 60 clinically relevant controls (Figure 1A), we performed a pilot study to assess the accuracy of 2 expert and 3 non-expert readers for the detection of CA using only A4C images.
Readers assessed all images in a fully-crossed design (Figure 1A) with and without the aid of the DL model output.
The reads consisted of binary interpretations indicating presence or absence of CA and high/low confidence in the interpretation.
Accuracy, sensitivity, and specificity were compared between aided and non-aided reads for statistical difference (paired t-test) and statistical equivalence (two paired one-sided t-tests).
Results
The DL model demonstrated an accuracy, sensitivity and specificity in the test dataset of 85% (95% CI: 81.6%, 88.2%), 88.3% (95% CI: 84.1, 92.3%), and 81.7% (95% CI 76.6, 80.3%), respectively (Figures 1B/1C).
Aided by the DL model, readers demonstrated the potential for small improvements in performance (Figure 1C/1D), with the suggestion of larger benefits to be observed in non-experts.
However, comparisons between aided and unaided reads were not significantly different, nor statistically equivalent, for any performance metric (all p ≥ 0.176).
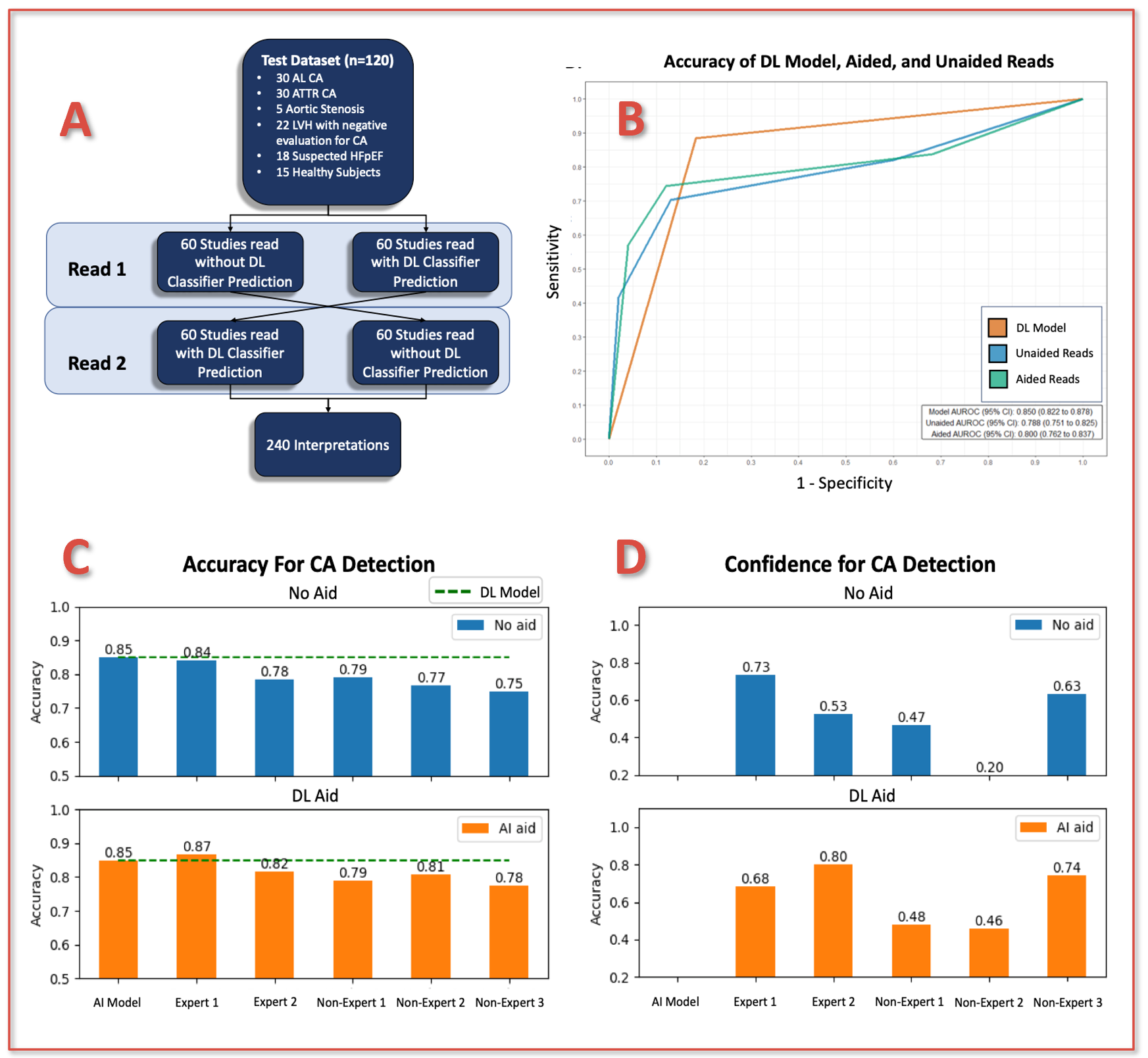
Figure 1. (A) Schematic of reader study design; (B) ROC curves for DL model, aided, and unaided readers in the test cohort. Comparison of reader (C) overall accuracy and (D) proportion of studies in which readers provided a confident interpretation for detection of CA with and without the aid of the DL model.
Conclusion
In this multicenter, multi-vendor study, we developed a novel DL model which demonstrated excellent performance for differentiating CA from clinically relevant controls.
Although the study demonstrates the potential for DL model to improve reader accuracy, particularly in non-experts, our findings indicate the need for a larger reader study to better understand if DL can augment clinical decision making to promote earlier CA diagnosis and treatment.
